Preparation of Cross-linked Enzyme Aggregates of Nitrile Hydratase ES-NHT-118 from E. coli by Macromolecular Cross-linking Agent
Liya Zhou, Haixia Mou, Jing Gao, Li Ma, Ying He and Yanjun Jiang
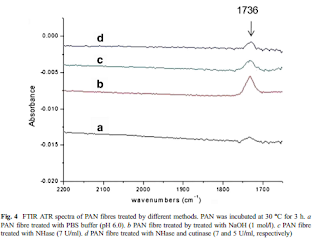
Cross-linked enzyme aggregates (CLEAs) of nitrile hydratase (NHase) ES-NHT-118 from E. coli were prepared by using ammonium sulfate as precipitating agent followed by cross-linking with dextran polyaldehyde for the first time. In this process, egg white was added as an amine source to aid formation of CLEAs. The optimal conditions of the immobilization process were determined. Michaelis constants (Km) of free NHase and NHase CLEAs were also determined. The NHase CLEAs exhibited increased stability at varied pH and temperature conditions compared to its free counterpart. When exposed to high concentrations of acrylamide, NHase CLEAs also exhibited effective catalytic activity.