The cobalt is no longer visible. However if you twirl the structure around you can find a view of it through a different aperture. I wonder how this impacts on the selectivity of this enzyme.
Showing posts with label PyMol. Show all posts
Showing posts with label PyMol. Show all posts
Wednesday, 6 July 2011
Comparing the access of the active site of 3QXE and 3HHT
If you orient the pdb file of the Geobacillus which is coded as 3HHT as I have done for 3QXE and 2QDY in the previous posts, you get the following two images.
Looking inside for the active site of 3QXE
This is just a couple of rough pictures of 3QXE (the nitrile hydratase from Pseudomonas putida), once again from PyMol, showing a ribbon view of it with the active site cobalt-binding residues shown as CPK spheres, followed by a surface model with some of the active site just visible inside. I have been careful to keep it oriented similarly to the AJ270 nitrile hydratase in the previous post.
Labels:
3QXE,
AJ270,
Pseudomonas putida,
PyMol,
Rhodoccocus erythropolis
Monday, 4 July 2011
Looking for the iron in AJ270
Using PyMol I have been looking to see what the tunnel which connects the iron centre to the outside world looks like. Here are some pictures. Iron is represented as a warmpink sphere, and the pdb structure used is 2QDY.
Or close up as...
Or close up as...
Wednesday, 9 February 2011
3D structure of AJ270 NHase
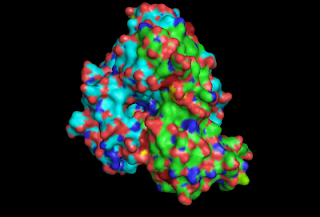
We have worked with the nitrile hydratase from Rhodococcus erythropolis AJ270 which was a bacterium which was collected on the banks of the river Tyne in Newcastle. The key authors who have worked with this bacterium in synthesis over the years are Otto Meth Cohn (at Sunderland University) and Mei-Xiang Wang (originally working with OMC but now at the Chinese Academy of Sciences, Beijing). M-X W has published a crystal structure of the nitrile hydratase from AJ270, and it can be found on PDB as 2QDY here. A little bit of playing with the data using PyMol and you can see how the two subunits fit together, and how there is a small channel from the exterior of the enzyme down into the active site.
Labels:
2QDY,
AJ270,
Meth Cohn,
nitrile hydratase,
PDB,
PyMol,
Rhodococcus,
Wang
Subscribe to:
Posts (Atom)